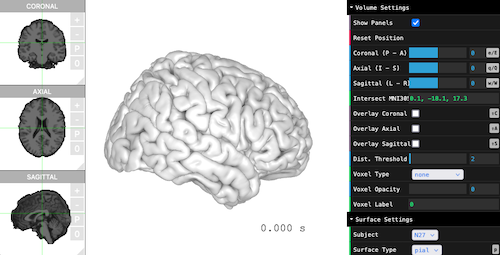
threeBrain is an R package that renders
FreeSurfer or AFNI/SUMA files in the modern
web browsers. The goal is to provide 3D viewers that are
powerful, interactive,
easy-to-share.
Directory Setup
The basic setup of a typical subject folder should look like this:
Project Directory
└─Subject code
├─mri - Atlas, T1 and/or CT image files (*.mgz, transforms, ...)
├─surf - FreeSurfer surface files (pial, sulc, ...)
├─SUMA - (Optional) SUMA standard 141 surfaces (std.141.*)
└─... - Other filesIf you are using FreeSurfer to generate MRI
reconstruction, then you are all set! For AFNI/SUMA users,
please place your SUMA folder under subject folder.
If you don’t have any FreeSurfer folders, you can
download a sample archive from here,
choose N27-complete.zip, and extract to your download
directory. Once extracted, you will have a N27 subject
folder in the ~/Downloads/ directory. The directory tree
looks exactly the same as described above.
In the following context, I will use ~/Downloads/N27 as
an example
Generate Viewer Object
library(threeBrain)
subject_code <- "N27"
subject_path <- "~/Downloads/N27"
brain <- threeBrain(subject_path, subject_code)
print(brain)
#> Subject - N27
#> Transforms:
#>
#> - FreeSurfer TalXFM [from scanner to MNI305]:
#> [,1] [,2] [,3] [,4]
#> [1,] 0.9692 -0.0029 -0.0134 -0.1638
#> [2,] 0.0062 0.9685 0.0492 -2.0717
#> [3,] 0.0145 0.0276 0.9541 0.1361
#> [4,] 0.0000 0.0000 0.0000 1.0000
#>
#> - Torig [Voxel CRS to FreeSurfer origin, vox2ras-tkr]
#> [,1] [,2] [,3] [,4]
#> [1,] -1 0 0 128
#> [2,] 0 0 1 -128
#> [3,] 0 -1 0 128
#> [4,] 0 0 0 1
#>
#> - Norig [Voxel CRS to Scanner center, vox2ras]
#> [,1] [,2] [,3] [,4]
#> [1,] -1 0 0 128.5
#> [2,] 0 0 1 -145.5
#> [3,] 0 -1 0 146.5
#> [4,] 0 0 0 1.0
#>
#> - Scanner center relative to FreeSurfer origin
#> [1] -0.5 17.5 -18.5
#>
#> - FreeSurfer RAS to MNI305, vox2vox-MNI305
#> [,1] [,2] [,3] [,4]
#> [1,] 0.9692 -0.0029 -0.0134 0.12365
#> [2,] 0.0062 0.9685 0.0492 -18.10715
#> [3,] 0.0145 0.0276 0.9541 17.31120
#> [4,] 0.0000 0.0000 0.0000 1.00000
#> Surface information (total count 1)
#> Loading required namespace: rstudioapi
#> pial [ std.141 ]
#> Volume information (total count 1)
#> T1If this is the first time, it might take a while to import and generate cache files.
Surface, Volume, Atlas
By default, the viewers only load the following information. Only the
pial surfaces are mandatory, other files are optional.
- T1 (MR images) and transforms (optional)
mri/brain.finalsurfs.mgzmri/transforms/talairach.xfm
-
pial(mandatory) andsulc(optional)-
surf/*h.pialorSUMA/std.141.*h.pial[.asc|gii] -
surf/*h.sulcorSUMA/std.141.*h.sulc.1D
-
-
aparc+aseg(optional)mri/aparc+aseg.mgz
The T1 image is automatically detected if
brain.finalsurfs.mgz is missing. The following alternatives
are brainmask.mgz, brainmask.auto.mgz,
T1.mgz.
To load more than one surfaces, please specify the surface types when loading the viewer. The available surface types are:
-
pial- (default) -
smoothwm,white- (smoothed) white matter -
inflated,sphere- inflated surfaces -
pial-outer-smoothed- (‘FreeSurfer’-only) smoothed surface tightly wrapping the pial surface -
inf_200- (‘SUMA’-only) inflated pial surface
The atlas type can be selected from aparc+aseg,
aparc.a2009s+aseg, aparc.DKTatlas+aseg, or
aseg. To load a specific atlas, please make sure the
corresponding file exists in mri/. For example,
mri/aseg.mgz.
The following example loads pial and
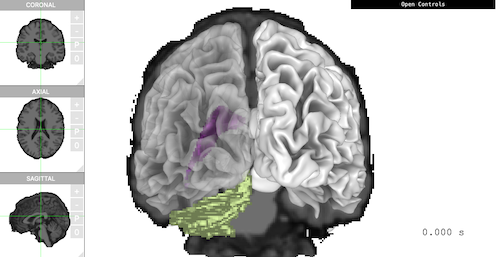
smoothwm, with aseg as atlas. The viewer shows
‘Coronal’ plane, smoothed white matter, ‘Ventricle’, and ‘Cerebellum’
all together in one scene.
brain <- threeBrain(
subject_path, subject_code,
surface_types = c('pial', 'smoothwm'),
atlas_types = 'aseg')
brain$plot(
controllers = list(
"Voxel Type" = "aseg",
"Voxel Label" = "4,5,6,7",
"Surface Type" = "smoothwm",
"Left Opacity" = 0.4,
"Overlay Coronal" = TRUE
),
control_display = FALSE,
camera_pos = c(0, -500, 0)
)