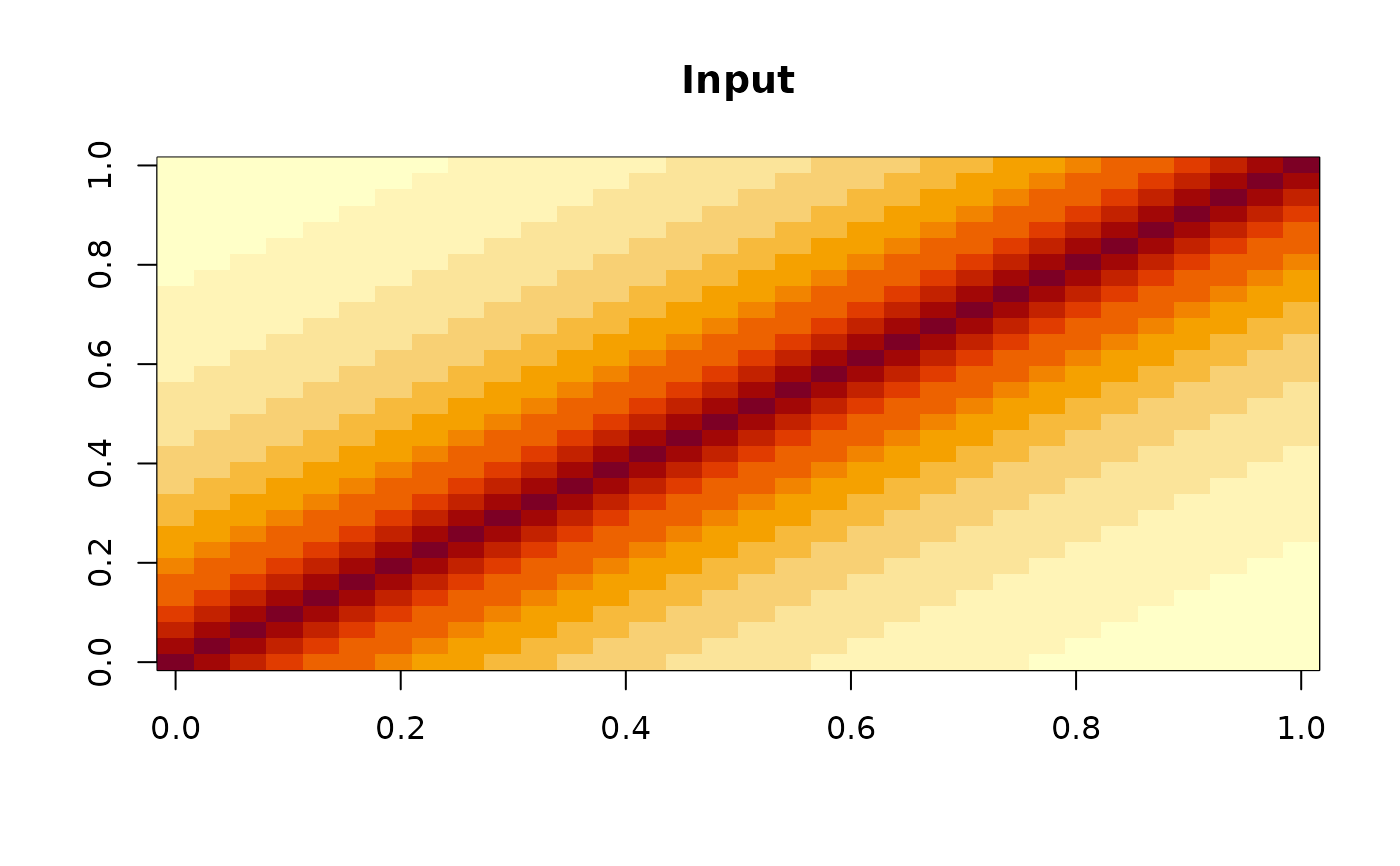
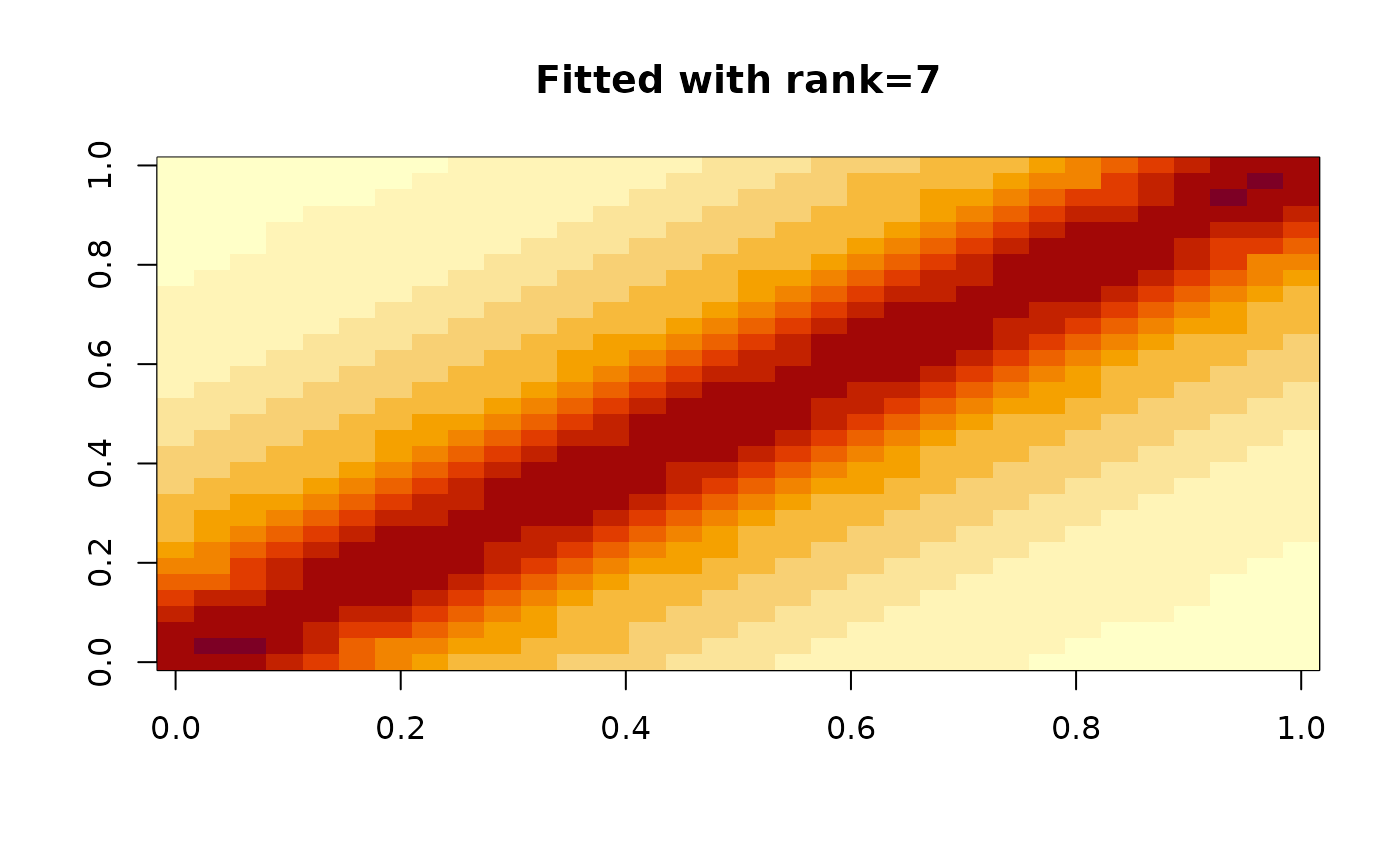
A pure-R vanilla implementation assuming inputs are non-negative matrices
without NA.
Usage
naive_nmf(x, k, tol = c(1e-04, 1e-08), max_iters = 10000, verbose = TRUE)Arguments
- x
a matrix, or can be converted into a matrix; all negative or missing values will be treated as zero
- k
decomposition rank
- tol
stop criteria, a numeric of two; the first number is the tolerance for root-mean-squared residuals, relative to the largest number in
x; the second number is the tolerance for weight differences; any stopping criteria met will result in the stop of iteration- max_iters
maximum iterations
- verbose
whether to report the progress; logical or a positive integer (of step intervals)